What is a transposable element (TE)?
Transposable elements are DNA sequences that can move from one location to another within the genome. Transposable elements (TEs), commonly known as jumping genes, were first discovered by Barbara McClintock in 1951 and won the Nobel Prize in 1983. Previously, scientists had rejected this discovery for many years and remained ignorant, but later proved through experiments that transposable factors are abundant in all organisms (prokaryotes and eukaryotes). They represent a large part of the genome, covering approximately 5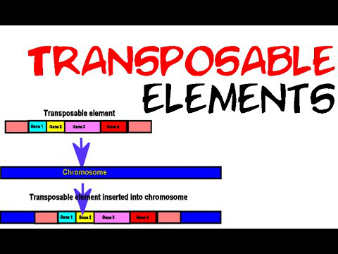
Types of transposable factors:
There are various types of TEs and different ways to classify them. However, the most common classification is based on transcription requirements. Type 1 or retrotransposons are those that require RNA to be transcribed into DNA for transposition, while type 2 or DNA transposons are transposons that do not require it for transposition.
Both Type 1 and Type 2 TE can be autonomous and involuntary in nature. Autonomous TEs can move on their own because they have the transposase or reverse transcriptase protein-coding genes required for transposition. Involuntary TEs cannot move on their own due to the lack of these proteins, so they rely on other TEs to borrow these proteins. The Ac element is autonomous, while the Ds element is essentially involuntary because they rely on Ac for transposition.
The role of the transposition element:
When TE covers 50% of the genome, a question arises in the mind, what do jumping genes do besides jumping? The jump-in of TE in another gene leads to mutations, for example, the insertion of L1 in the factor VIII gene leads to hemophilia. The researchers also found that the APC gene in colon cancer cells has an L1 transposition that is not found in normal cells. This shows the role of TE in disease development.
Except for L1, TE usually remains silent most of the time. Some of them remain silent because their mutations inactivate them and hinder their ability to move around chromosomal locations, while others are prevented by epigenetic factors such as chromatin remodeling, DNA methylation, or miRNA. For example, chromatin remodeling prevents certain regions from packaging transcriptase in a contracted manner, thereby silencing TE in these regions.
The movement of TE does not always cause harm. They can also play a key role in genome evolution by shuffling exons, transferring genes, and repairing DNA breaks. For example, the ToI2 DNA transposon determines the pigmentation of medaka with multiple pigmentation patterns. It was found that when the ToI2 transposon jumped out without disturbing the genome, the fish became albino, and when they were present, the fish could acquire a wide range of heritable pigmentation patterns





